Abstract
Introduction: New Diffuse Large B-cell (DLBCL) treatments remain a clinical need despite the success of rituximab combined with CHOP chemotherapy (RCHOP), which results in durable responses in 60-70% of patients. Those refractory to or who relapse following first-line therapy (rrDLBCL) have a very poor outcome, with only 20% surviving beyond 5 years, although CAR-T cell therapy is making a significant impact. Genomic insights can inform mechanism and treatment studies within this population, specifically the precision application of CAR-T. Rushton et. al. recently analyzed a large rrDLBCL patient cohort, noting significant rises in specific genes during the transition to advanced disease (MS4A1, TP53, NFKBIE, FOXO1, and CREBBP), building on the innovations of pre-treatment classifications. Addressing the heterogeneous activation of oncogenic pathways remains a challenge. Inhibitors targeting genes responsible for epigenetic regulation of rrDLBCL represent an attractive option, namely the transcriptional regulator BRD4, with PROTAC inhibition resulting in downregulation of several key DLBCL oncogenes. Herein, we report the first genomic subclassifications of rrDLBCL, the enrichment of TP53 alterations in the EZB-like subclassification, and data supporting the application of BRD4-targeting PROTAC therapies.
Methods: Non-negative matrix factorization (NMF) was applied to a publicly available cohort of 127 RCHOP-treated rrDLBCL patients with matching sequencing data. Marker selection was applied to the 2 emergent groups to detect differential presence of DNA alterations. A Pearson similarity matrix was applied to the rrDLBCL cohort and 3 pretreatment cohorts to detect patterns of DNA alteration association and exclusion. Next, CRISPR-KO gene scores were integrated against gene expression fold change values before and after BRD4 PROTAC treatment. Lasty, 4 DLBCL cell lines were assayed against the BRD4 PROTAC ARV-825, Venetoclax, and inhibitors of epigenetic regulation.
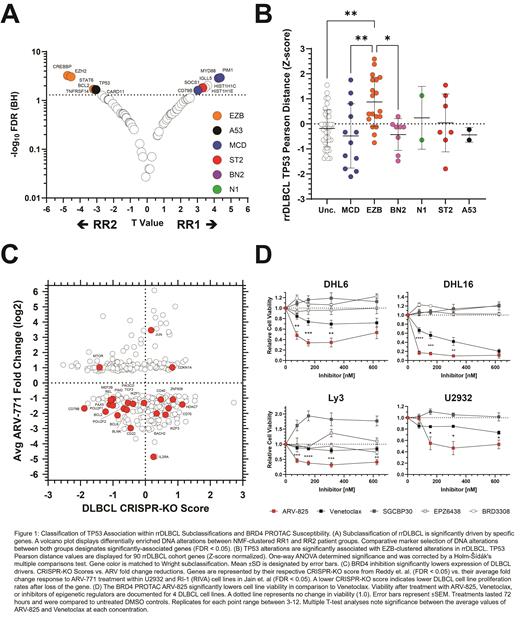
Results: NMF produced 2 clusters based on DNA alterations from the rrDLBCL cohort: RR1 and RR2. Significantly associated genes (FDR ≤ 0.05) included MYD88, PIM1, IGLL5, HIST1HC, SOCS1, HIST1H1E, and CD79B in RR1 , and CREBBP, EZH2, STAT6, BCL2, TP53, TNFRSF14, CARD11, and double hit translocations in RR2. Notably, TP53 alterations significantly enriched in the EZB subclassification during rrDLBCL transition in comparison to pre-treatment distribution (P = 0.0016). Significant associations between TP53 and EZB genes were observed alongside significant exclusion from MCD and BN2-associated genes. Integrating significant gene expression changes following BRD4-PROTAC treatment and CRISPR-KO scores revealed the loss of several key oncogenes in cell lines, including BCL2, BCL6, CD79B, POU2F2, and MDM4. Markedly, reductions in 9 established GCB and 4 ABC gene signatures were observed alongside the reduction of the Reactome signature of TP53 Regulators of Expression and Degradation (FDR = 0.0377). In vitro inhibition assays measuring ARV-825 (BRD4 PROTAC) response revealed significantly lower rates of cell viability compared to Venetoclax across 4 cell lines.
Conclusions: Our results indicate that distinct genomic groups exist within rrDLBCL, that TP53 alterations strongly associate with the EZB-enriched group, and that if TP53 alterations continue to predict poor CAR-T response, these tumors may be susceptible to BRD4 inhibition. Our observations indicate a significant climb in TP53 association with EZB tumors and their matched alterations after rrDLBCL transition in comparison to the pre-treatment landscape. Enrichment may indicate that EZB-like tumors lack some of the tools necessary to make the transition to advanced disease but provide an ideal environment for the acquisition of TP53 alterations. In contrast, the disassociation between TP53 alterations and MCD/BN2-like alterations may indicate that these tumors have more inherent capability to reject therapy. The emergence of BRD4-targeting PROTACs provides an additional clinical tool, one perhaps more suitable than Venetoclax against specific rrDLBCL cases. In closing, This study provides insights towards the rrDLBCL landscape and reinforces the necessity of evaluating tissue biopsies for TP53 alterations throughout treatment to guide precision application of therapy, noting susceptible pathways of these particular tumors.
No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal